Phylogenetic & Network Methods
Tree inference, placement, and network analysis.

T-BAS Toolkit
Tree-Based Alignment Selector Toolkit.
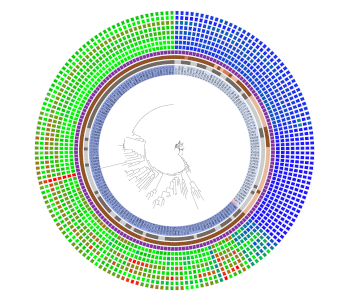
Patristic Distance
Create patristic distance rings with DendroPy.
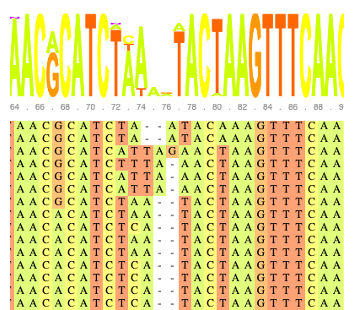
MAFFT
Align sequences with MAFFT.
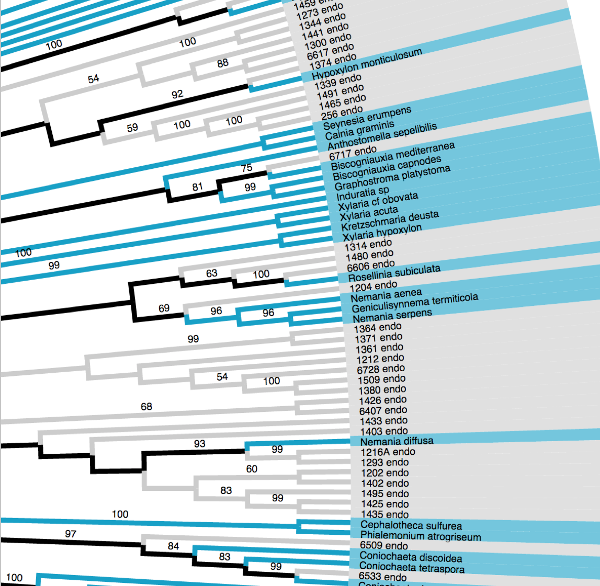
RAxML & FastTree
Maximum likelihood tree inference.
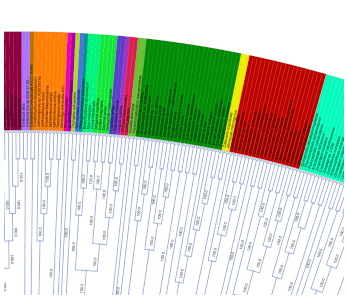
IQ-TREE
Fast maximum likelihood tree inference.
BEAST
Bayesian phylogenetic tree inference.
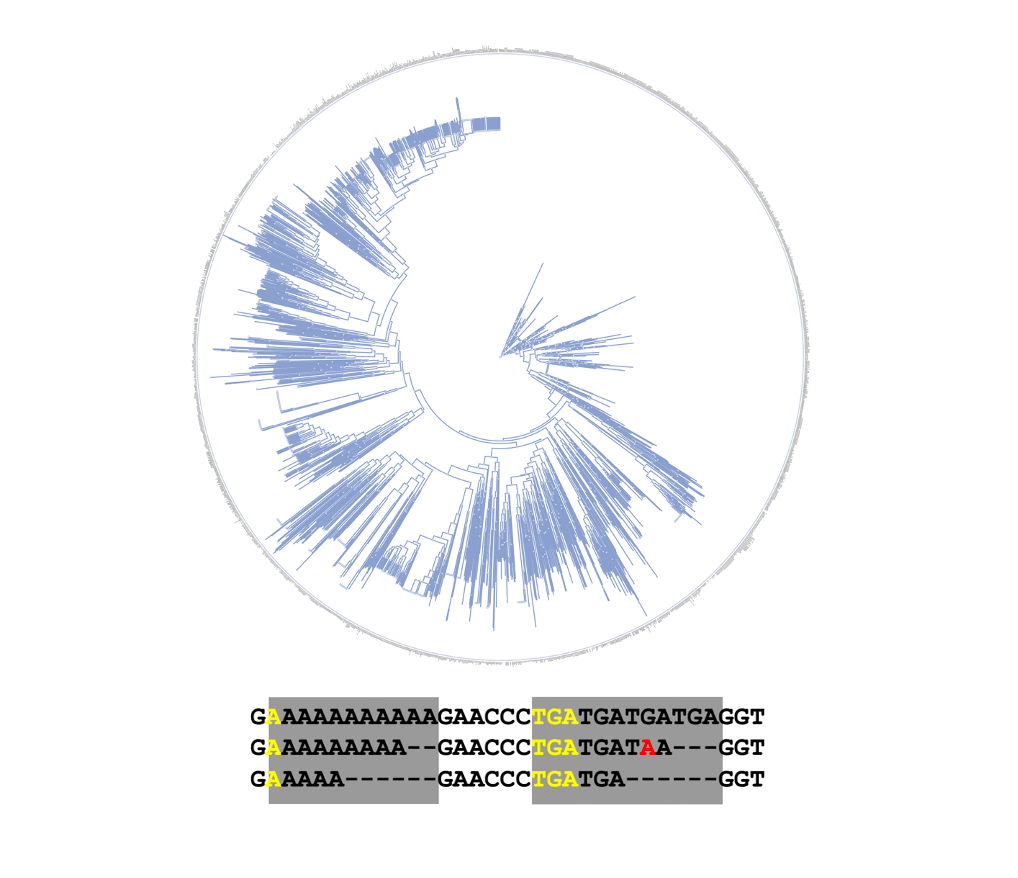
Microsatellite Tree
Reconstruct tree from microsatellite data.
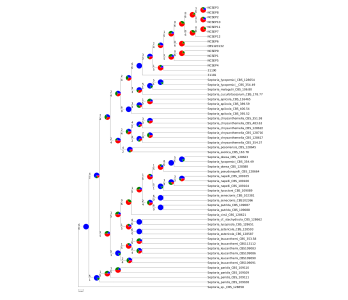
ASTRAL
Multispecies coalescent; PhyParts gene-tree conflict.
EPA-ng
Parallel, highly accurate ML phylogenetic placement.
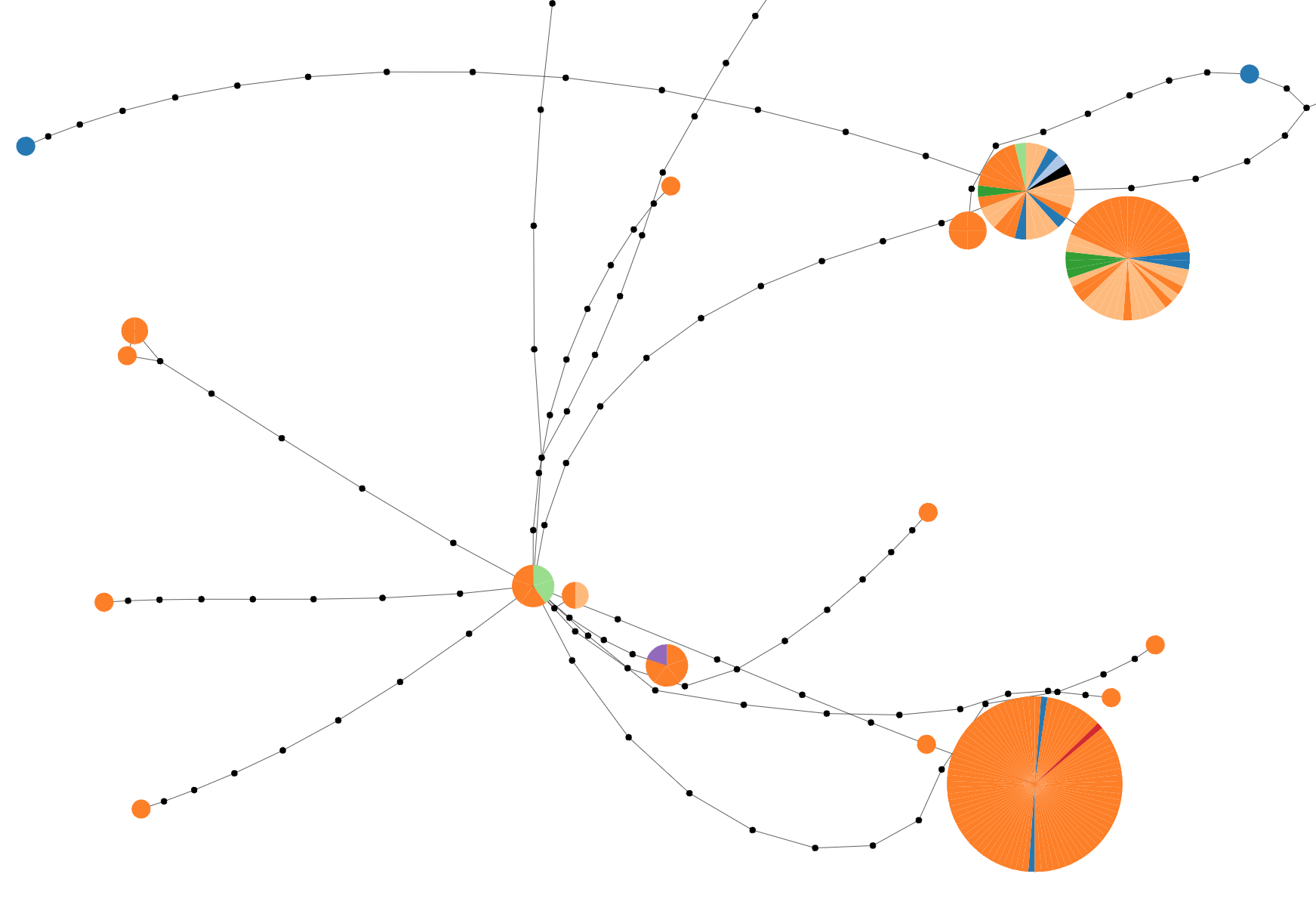
Graph Inference
Create networks with TCS or upload graph file.
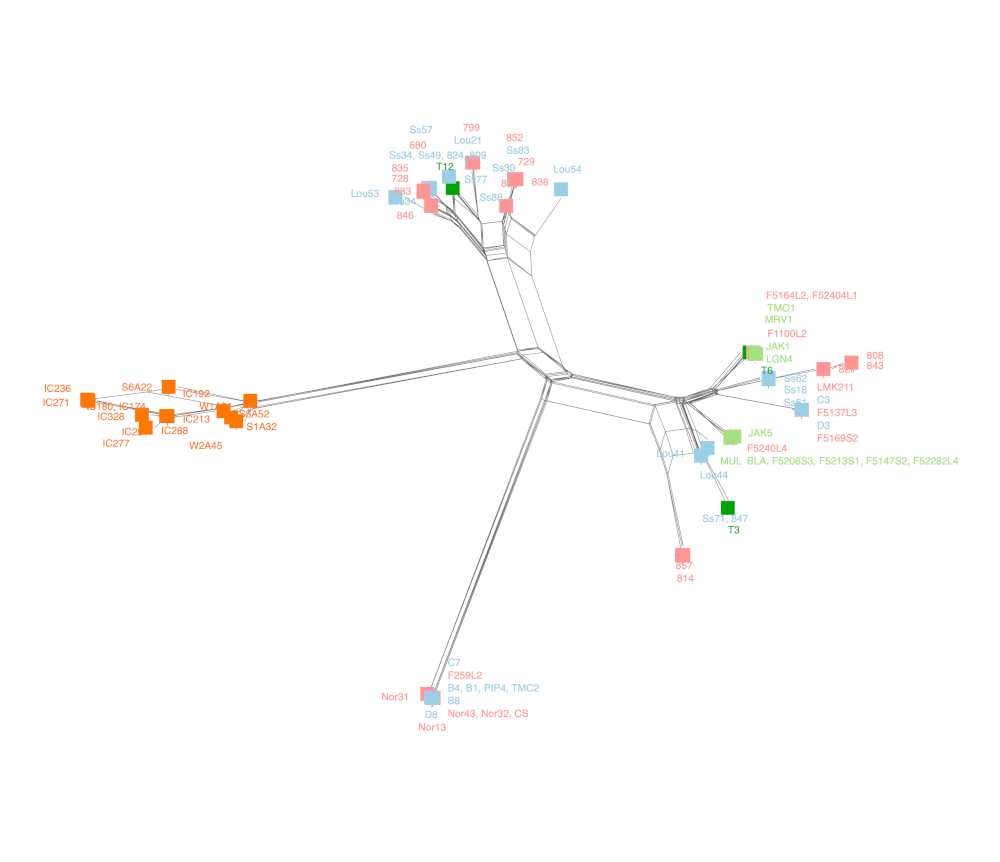
SplitsTree
Neighbor-Net splits network inference.

Mesquite Hypha
Display multiple node support values on trees.